Building an accurate model of the SARS-CoV-2 virus
23/12/2020 UPDATE - Please scroll to the bottom of this page for the new model!
On 23 March 2020 the UK entered national lockdown. Prime Minister Boris Johnson appeared on TV to order everyone to stay at home, looking uncharacteristically serious.
At this time I was living in Glasgow and waiting to begin my next job as Digital Media Specialist for the International Livestock Research Institute. Like everyone else, I was also thinking a lot about SARS-CoV-2, COVID-19, and viruses.
I decided my free time could be usefully spent building an accurately proportioned model of the SARS-CoV-2 virus. I created it using information from current published research, using 3Ds Max 2020. I contacted Dr Edward Hutchinson from the MRC-Centre for Virus Research, University of Glasgow, for advice, and he helpfully provided feedback and recommendations of which science papers to use.
From my MSc in Medical Visualisation I knew the rudiments of how to use 3Ds Max. However, the course focused purely on depicting anatomy, not molecular or cellular biology. This means I needed to work out how to model the virus myself, through researching techniques online.
I had also worked as a research assistant at the School of Life Sciences, University of Glasgow, with Dr Craig Daly, senior lecturer in physiology. Together we had devised ways of converting confocal microscopy data into digital 3D models. This work had also introduced me to Sketchfab, a useful and publically available platform for showcasing digital models. I reasoned that if I could create an accurate 3D model of the SARS-CoV-2 coronavirus, I should put it on Sketchfab to make it widely viewable and shareable.
This would, however, mean that the model would need to fall under a certain size limit, and is therefore why I chose to take certain artistic licences and approximations in its construction, which are detailed below.
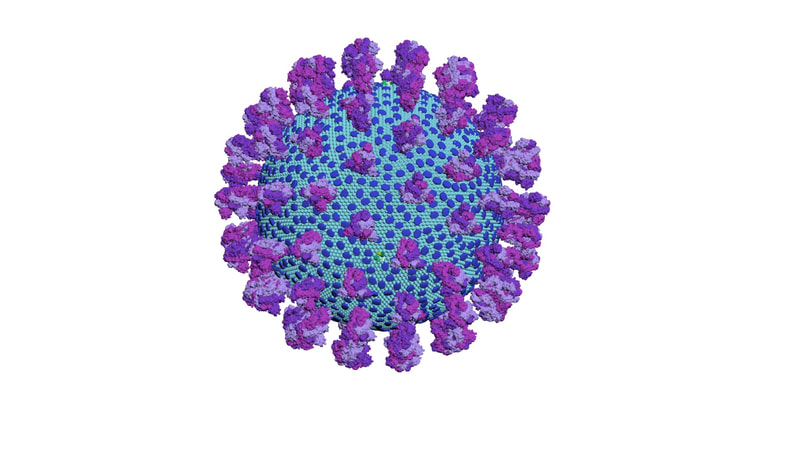
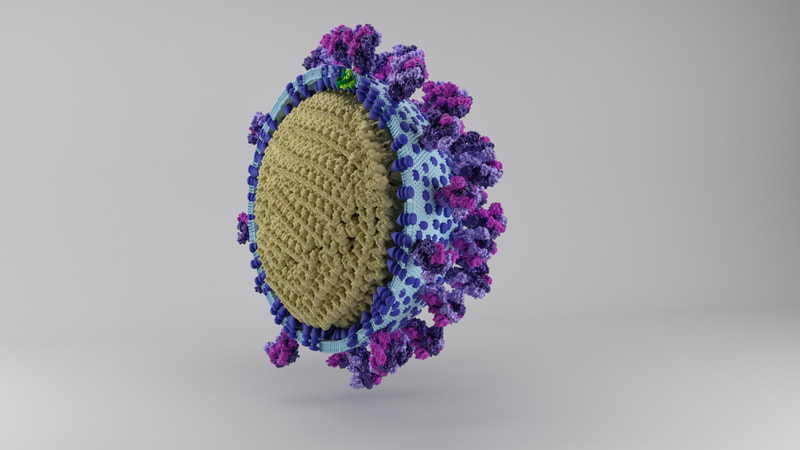
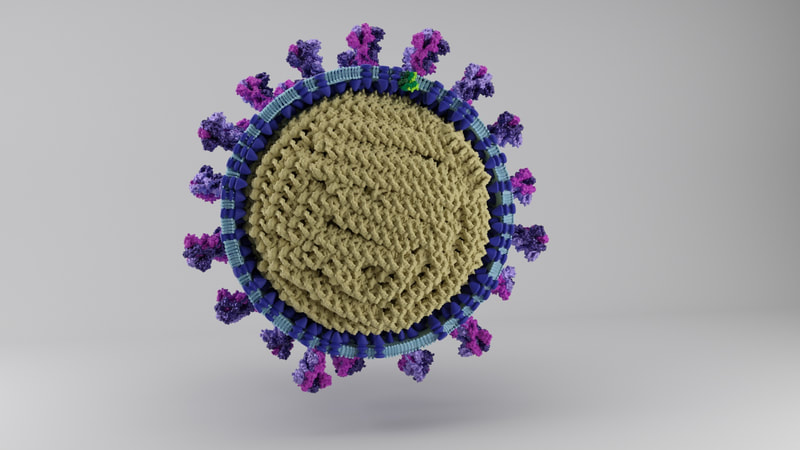
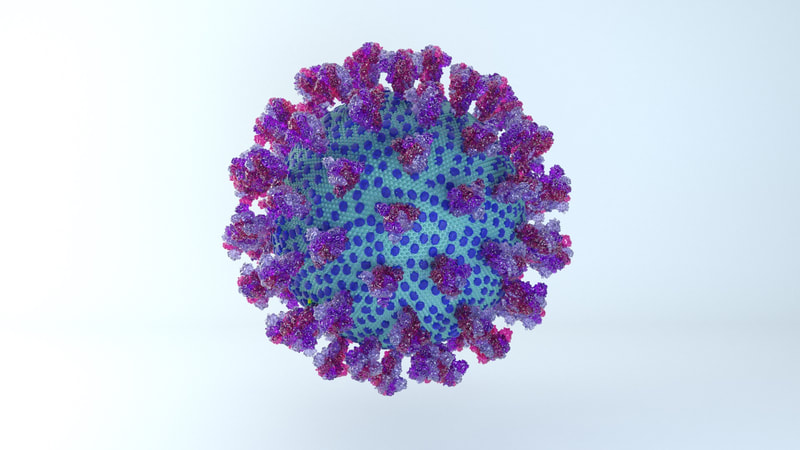
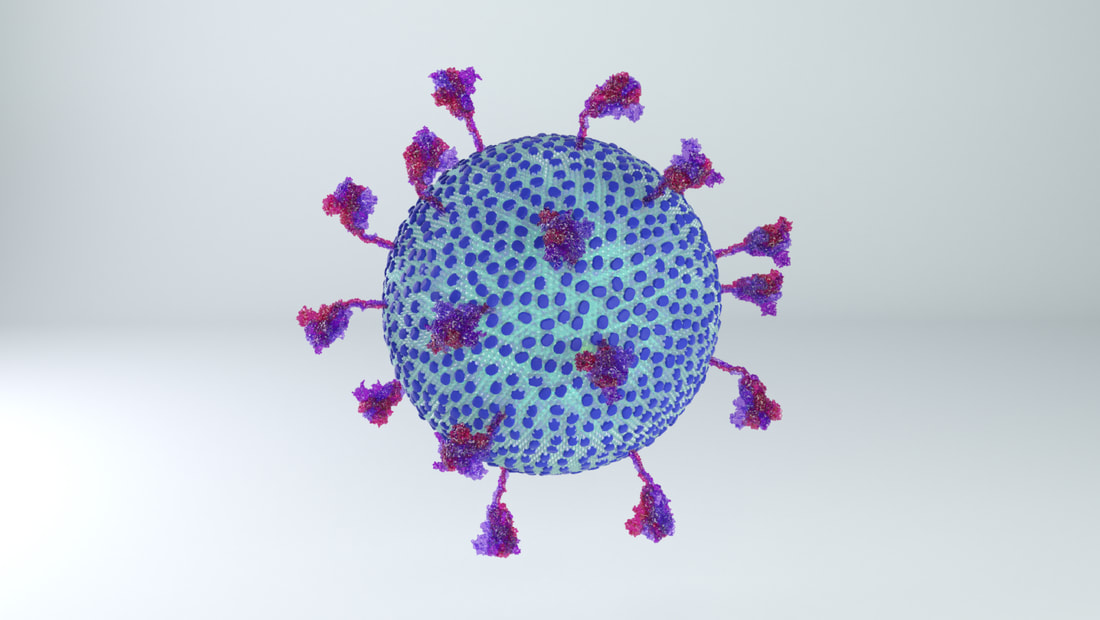
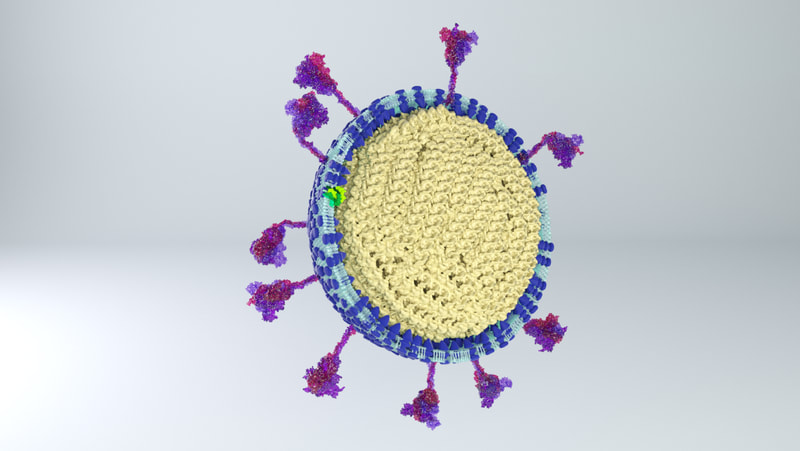
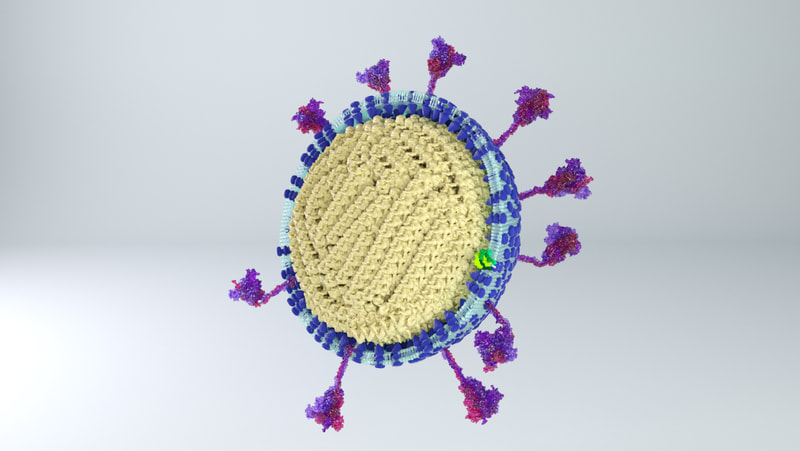
I constructed three model designs - a section of the virus envelope, the whole virion, and a half-section view of the virion. These were published on 27 May 2020 and promoted by local and university press.
I'm happy to credit Dr Hutchinson for his help, and the University of Glasgow and Glasgow School of Art for helping to promote the models. I would like to stress though that these models are my voluntary work, unpaid, done in the evenings and weekends alongside my day job, over a couple of months. I also had to use my own equipment and until I had my first paycheque from my day job and could buy a new desktop computer, I was rendering views of the model by balancing an ageing laptop on top of trays of ice to keep it from overheating. All quite the challenge!
What would really make me happy would be if I could take these models further. Without file size for upload being a constraint, there's a lot that could be done to make them more detailed, updated, or nicely animated. It would also be a lot quicker than the first time round! I hope I can be hired by someone to do so.
On 23 March 2020 the UK entered national lockdown. Prime Minister Boris Johnson appeared on TV to order everyone to stay at home, looking uncharacteristically serious.
At this time I was living in Glasgow and waiting to begin my next job as Digital Media Specialist for the International Livestock Research Institute. Like everyone else, I was also thinking a lot about SARS-CoV-2, COVID-19, and viruses.
I decided my free time could be usefully spent building an accurately proportioned model of the SARS-CoV-2 virus. I created it using information from current published research, using 3Ds Max 2020. I contacted Dr Edward Hutchinson from the MRC-Centre for Virus Research, University of Glasgow, for advice, and he helpfully provided feedback and recommendations of which science papers to use.
From my MSc in Medical Visualisation I knew the rudiments of how to use 3Ds Max. However, the course focused purely on depicting anatomy, not molecular or cellular biology. This means I needed to work out how to model the virus myself, through researching techniques online.
I had also worked as a research assistant at the School of Life Sciences, University of Glasgow, with Dr Craig Daly, senior lecturer in physiology. Together we had devised ways of converting confocal microscopy data into digital 3D models. This work had also introduced me to Sketchfab, a useful and publically available platform for showcasing digital models. I reasoned that if I could create an accurate 3D model of the SARS-CoV-2 coronavirus, I should put it on Sketchfab to make it widely viewable and shareable.
This would, however, mean that the model would need to fall under a certain size limit, and is therefore why I chose to take certain artistic licences and approximations in its construction, which are detailed below.
I constructed three model designs - a section of the virus envelope, the whole virion, and a half-section view of the virion. These were published on 27 May 2020 and promoted by local and university press.
I'm happy to credit Dr Hutchinson for his help, and the University of Glasgow and Glasgow School of Art for helping to promote the models. I would like to stress though that these models are my voluntary work, unpaid, done in the evenings and weekends alongside my day job, over a couple of months. I also had to use my own equipment and until I had my first paycheque from my day job and could buy a new desktop computer, I was rendering views of the model by balancing an ageing laptop on top of trays of ice to keep it from overheating. All quite the challenge!
What would really make me happy would be if I could take these models further. Without file size for upload being a constraint, there's a lot that could be done to make them more detailed, updated, or nicely animated. It would also be a lot quicker than the first time round! I hope I can be hired by someone to do so.
How I did it
References:
At the time of construction (March - May 2020) I used measurements from studies about the SARS-CoV-2 virion, and from studies of similar coronaviruses. These include the coronaviruses responsible for SARS and for mouse hepatitis.
I used the following scientific papers to find proportions for the virion elements:
Barcena, M., Oostergetel, G., Bartelink, W., Faas, F., Verkleij, A., & Rottier, P. et al. (2009). Cryo-electron tomography of mouse hepatitis virus: Insights into the structure of the coronavirion. Proceedings Of The National Academy Of Sciences, 106(2), 582-587. doi: 10.1073/pnas.0805270106
Beniac, D., Andonov, A., Grudeski, E., & Booth, T. (2006). Architecture of the SARS coronavirus prefusion spike. Nature Structural & Molecular Biology, 13(8), 751-752. doi: 10.1038/nsmb1123
Chang, C., Hou, M., Chang, C., Hsiao, C., & Huang, T. (2014). The SARS coronavirus nucleocapsid protein – Forms and functions. Antiviral Research, 103, 39-50. doi: 10.1016/j.antiviral.2013.12.009
Jayaram, H., Fan, H., Bowman, B., Ooi, A., Jayaram, J., & Collisson, E. et al. (2006). X-Ray Structures of the N- and C-Terminal Domains of a Coronavirus Nucleocapsid Protein: Implications for Nucleocapsid Formation. Journal Of Virology, 80(13), 6612-6620. doi: 10.1128/jvi.00157-06
Neuman, B., Adair, B., Yoshioka, C., Quispe, J., Orca, G., & Kuhn, P. et al. (2006). Supramolecular Architecture of Severe Acute Respiratory Syndrome Coronavirus Revealed by Electron Cryomicroscopy. Journal Of Virology, 80(16), 7918-7928. doi: 10.1128/jvi.00645-06
And a section of this textbook:
Fields, B., Knipe, D., & Howley, P. (2013). Fields virology. Philadelphia: Wolters Kluwer/Lippincott Williams & Wilkins Health.
I used the following scientific papers to find proportions for the virion elements:
Barcena, M., Oostergetel, G., Bartelink, W., Faas, F., Verkleij, A., & Rottier, P. et al. (2009). Cryo-electron tomography of mouse hepatitis virus: Insights into the structure of the coronavirion. Proceedings Of The National Academy Of Sciences, 106(2), 582-587. doi: 10.1073/pnas.0805270106
Beniac, D., Andonov, A., Grudeski, E., & Booth, T. (2006). Architecture of the SARS coronavirus prefusion spike. Nature Structural & Molecular Biology, 13(8), 751-752. doi: 10.1038/nsmb1123
Chang, C., Hou, M., Chang, C., Hsiao, C., & Huang, T. (2014). The SARS coronavirus nucleocapsid protein – Forms and functions. Antiviral Research, 103, 39-50. doi: 10.1016/j.antiviral.2013.12.009
Jayaram, H., Fan, H., Bowman, B., Ooi, A., Jayaram, J., & Collisson, E. et al. (2006). X-Ray Structures of the N- and C-Terminal Domains of a Coronavirus Nucleocapsid Protein: Implications for Nucleocapsid Formation. Journal Of Virology, 80(13), 6612-6620. doi: 10.1128/jvi.00157-06
Neuman, B., Adair, B., Yoshioka, C., Quispe, J., Orca, G., & Kuhn, P. et al. (2006). Supramolecular Architecture of Severe Acute Respiratory Syndrome Coronavirus Revealed by Electron Cryomicroscopy. Journal Of Virology, 80(16), 7918-7928. doi: 10.1128/jvi.00645-06
And a section of this textbook:
Fields, B., Knipe, D., & Howley, P. (2013). Fields virology. Philadelphia: Wolters Kluwer/Lippincott Williams & Wilkins Health.
An outline:
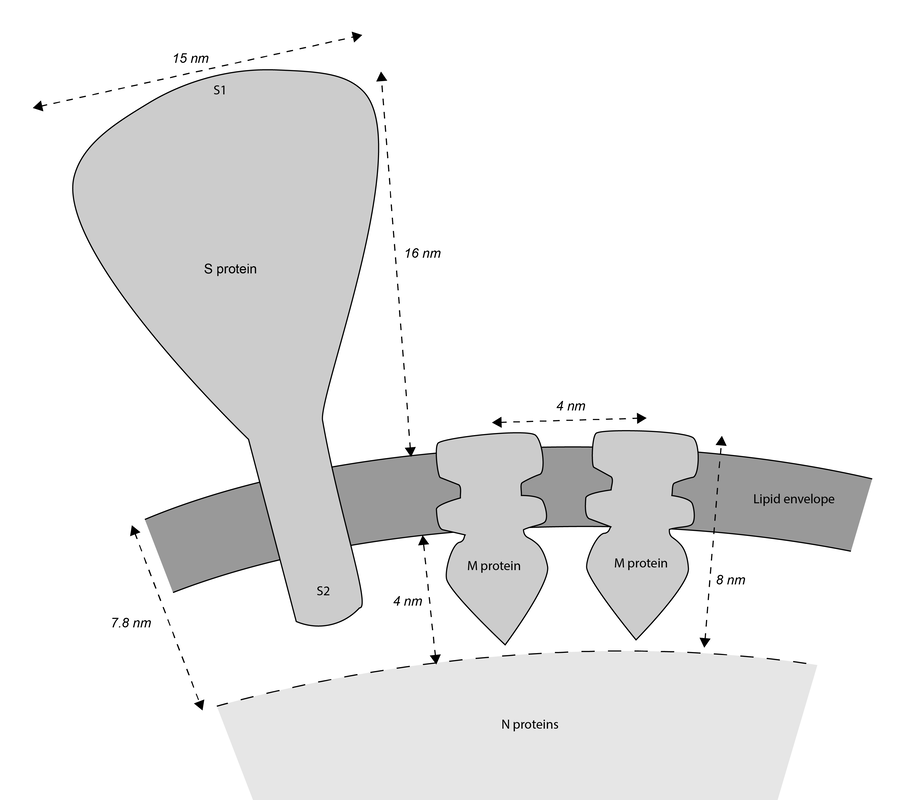
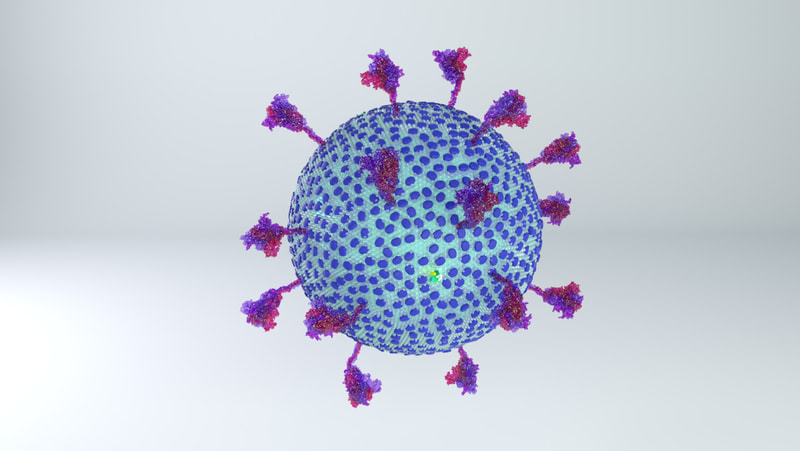
Coronaviruses are rounded shapes with a corona of projecting spikes. Beniac et al (2006) describe the SARS coronavirus as having a diameter of 118.5nm, including spikes, and the spikes themselves being 16 nm in height.
With a touch of rounding, this means the full diameter including spikes would be 120 nm, and without spikes - the lipid membrane and everything else in it - should be 88 nm in diameter.
With a touch of rounding, this means the full diameter including spikes would be 120 nm, and without spikes - the lipid membrane and everything else in it - should be 88 nm in diameter.
|
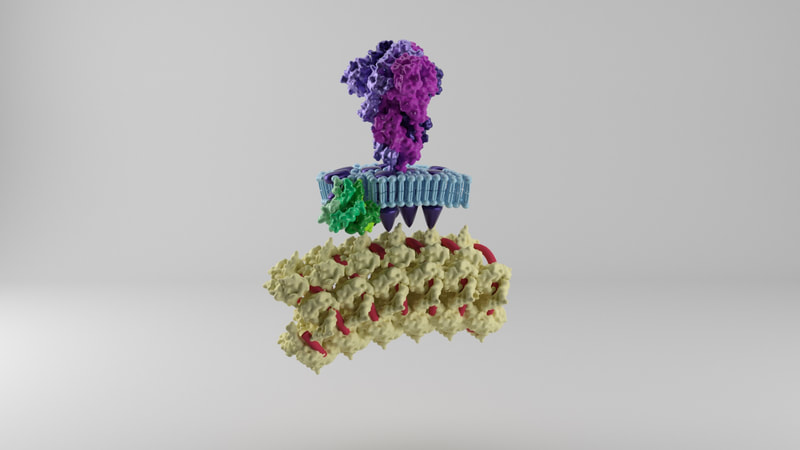
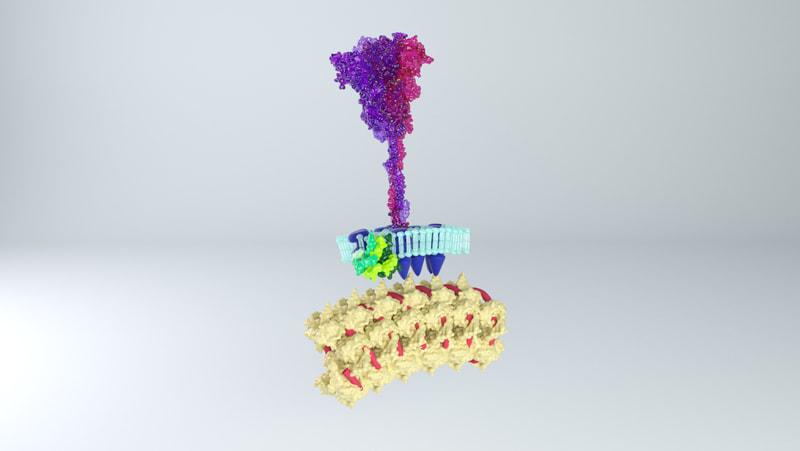
Spike proteins
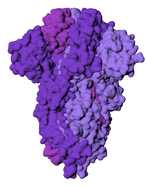
While I was crafting the model, 3D models of the spikes for SARS-CoV-2 became available on the Protein Data Bank. How many spikes? Anywhere from 60 - 100, suggested Beniac et al, based on the SARS coronavirus, of 16 nm in height, and 15 nm in width. |
|
Membrane proteins
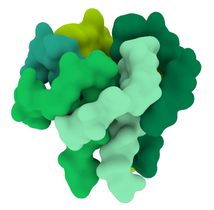
The lipid membrane of the virus is also studded through with membrane proteins and envelope proteins. Neuman et al (2011) say the SARS coronavirus membrane is about 4 nm thick. The membrane proteins fall into two forms - compact and long. I opted for the long form, described as being 8 nm in length, spaced around 4 nm apart. The authors describe the M protein as having a 'dagger-shape'. There was an approximation of the shape of SARS-CoV-2 M protein, worked out through a protein folding modelling program by the Zhang Lab. On the other hand, the shape did not resemble the shape described by Neuman et al, and would not be easy to arrange proportionately in the model. I opted to model a dagger-shape structure and use that instead. How many M proteins? According to Neuman et al, around 1100 on average. |
|
Envelope proteins
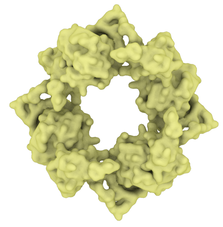
Fields Virology describe there being a copy number of about 20 envelope proteins in a coronavirus. Envelope proteins form pentameric cone-like structures, meaning there would be four assemblies in the virus envelope, of a size able to cross the 4nm lipid membrane. Again, a structure for the SARS coronavirus was available on the Protein Data Bank. |
|
Nucleoprotein and RNA
Inside the virus would be a core of coiled nucleoprotein and RNA. Bárcena et al (2009) and Chang et al (2014) describe the nucleoprotein as forming a well packed, hollow supercoil of interior diameter 3-4 nm, exterior diameter 9-16 nm. Neuman et al (2011) also describe a space around the exterior of the nucleoprotein core, partly filled by the tail ends of the membrane proteins which interact with the nucleoprotein. |
While I've included the best references above for each element size and statistic, generally all figures were agreed upon across multiple references statistics. All in all, they would result in these proportions:
Putting it together
3Ds Max provides grid and measurement tools to allow accurate design and creation. I chose a scale and stuck to it, checking the sizes of each element in proportion to each other.
My first challenge came by trying to think how to make a 3D membrane that would incorporate many different elements, without them overlapping, of a correct distance from each other - say, 90 spike proteins positioned equidistant from each other, but also equidistant from the 1100-odd membrane proteins and five pentameric envelope proteins assortments. Neuman et al (2011) also stated that each spike protein tended to be arranged over a 'rhombus' of 8 membrane proteins.
I tried, and I tried. I asked, I googled, I experimented. I could figure out how to make multiple spherical arrangements of each separate element, but if I put them together, inevitably they would overlap, or not be equally spaced from each other. This would be okay for most models or animations, but I was committed to being scientifically accurate.
I suspect any seasoned 3Ds Max artist who comes across this article will know a comparatively simple way of doing this. Or, perhaps, any 3D modeller with an immensely powerful computer that can run the processing to sort it out. In the end, I largely did it using certain 3Ds Max modifiers, calculations, and a LOT of manual adding, rotating and nudging around of elements. I found the best proportion of membrane proteins and spike proteins to fall within the ranges and distances described in the scientific literature, and the capabilities of the 3Ds Max tools I used.
If I had to do them again, I might reduce the number of M proteins (currently at around 1200) in order to create more distance between them. They measure at 4 nm in distance from each other if measured from the centre of each protein. It is possible that they should be further apart.
But the world of microbiology is a messy, organic place, and while viruses are highly ordered structures, even they would have a little variation in size, shape and distance here and there. After all the SARS-CoV-2 virus is not actually a perfect sphere in the first place.
I also simplified the lipid membrane. A lipid membrane is a close-packed layer of bobbing phospholipids. For simplicity and small file size, I constructed a membrane made out of two dimpled spheres, instead of many tiny elements. Shame though - there are some really cool animations that can be made of membranes disintegrating and flying apart!
My first challenge came by trying to think how to make a 3D membrane that would incorporate many different elements, without them overlapping, of a correct distance from each other - say, 90 spike proteins positioned equidistant from each other, but also equidistant from the 1100-odd membrane proteins and five pentameric envelope proteins assortments. Neuman et al (2011) also stated that each spike protein tended to be arranged over a 'rhombus' of 8 membrane proteins.
I tried, and I tried. I asked, I googled, I experimented. I could figure out how to make multiple spherical arrangements of each separate element, but if I put them together, inevitably they would overlap, or not be equally spaced from each other. This would be okay for most models or animations, but I was committed to being scientifically accurate.
I suspect any seasoned 3Ds Max artist who comes across this article will know a comparatively simple way of doing this. Or, perhaps, any 3D modeller with an immensely powerful computer that can run the processing to sort it out. In the end, I largely did it using certain 3Ds Max modifiers, calculations, and a LOT of manual adding, rotating and nudging around of elements. I found the best proportion of membrane proteins and spike proteins to fall within the ranges and distances described in the scientific literature, and the capabilities of the 3Ds Max tools I used.
If I had to do them again, I might reduce the number of M proteins (currently at around 1200) in order to create more distance between them. They measure at 4 nm in distance from each other if measured from the centre of each protein. It is possible that they should be further apart.
But the world of microbiology is a messy, organic place, and while viruses are highly ordered structures, even they would have a little variation in size, shape and distance here and there. After all the SARS-CoV-2 virus is not actually a perfect sphere in the first place.
I also simplified the lipid membrane. A lipid membrane is a close-packed layer of bobbing phospholipids. For simplicity and small file size, I constructed a membrane made out of two dimpled spheres, instead of many tiny elements. Shame though - there are some really cool animations that can be made of membranes disintegrating and flying apart!
Results
Where are they now?
Three models are on Sketchfab, also with labelling and additional information. Like I say, because of the constraints of file size, there's a couple of caveats. The whole virus and half virus models are actually hollow, instead of packed chock-full of nucleoprotein. The RNA is not visible on the nucleoprotein in these models, both because Dr Hutchinson advised it would be pretty tiny anyway, and more because I hadn't yet come up with a way to model one long coil of nucleoprotein and RNA. And, to make the files smaller, I used mesh reduction tools and programs, which have softened the resolution a little.
But they're proportionally accurate, based on the research at the time!
But they're proportionally accurate, based on the research at the time!
Here's the video version of the above!
23/12/20
And now, an updated model
This is the most recent update of my SARS-CoV-2 model, based on the latest research.
References:
The new model is based on the findings described in the following papers:
Ke, Z., Oton, J., Qu, K. et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature 588, 498–502 (2020). https://doi.org/10.1038/s41586-020-2665-2
Yao, H., Song, Y., Chen, Y., Wu, N., Xu, J., & Sun, C. et al. (2020). Molecular Architecture of the SARS-CoV-2 Virus. Cell, 183(3), 730-738.e13. doi: 10.1016/j.cell.2020.09.018
I am using a new model for the spike protein, provided by this research group:
Casalino, L., Gaieb, Z., Goldsmith, J., Hjorth, C., Dommer, A., & Harbison, A. et al. (2020). Beyond Shielding: The Roles of Glycans in the SARS-CoV-2 Spike Protein. ACS Central Science, 6(10), 1722-1734. doi: 10.1021/acscentsci.0c01056
Ke, Z., Oton, J., Qu, K. et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature 588, 498–502 (2020). https://doi.org/10.1038/s41586-020-2665-2
Yao, H., Song, Y., Chen, Y., Wu, N., Xu, J., & Sun, C. et al. (2020). Molecular Architecture of the SARS-CoV-2 Virus. Cell, 183(3), 730-738.e13. doi: 10.1016/j.cell.2020.09.018
I am using a new model for the spike protein, provided by this research group:
Casalino, L., Gaieb, Z., Goldsmith, J., Hjorth, C., Dommer, A., & Harbison, A. et al. (2020). Beyond Shielding: The Roles of Glycans in the SARS-CoV-2 Spike Protein. ACS Central Science, 6(10), 1722-1734. doi: 10.1021/acscentsci.0c01056
So what's changed?
A lot! I've incorporated some of the main changes into the model so that the appearance is much more accurate. Of course, there's still plenty of fine-tuning that could be done, and perhaps will be done.
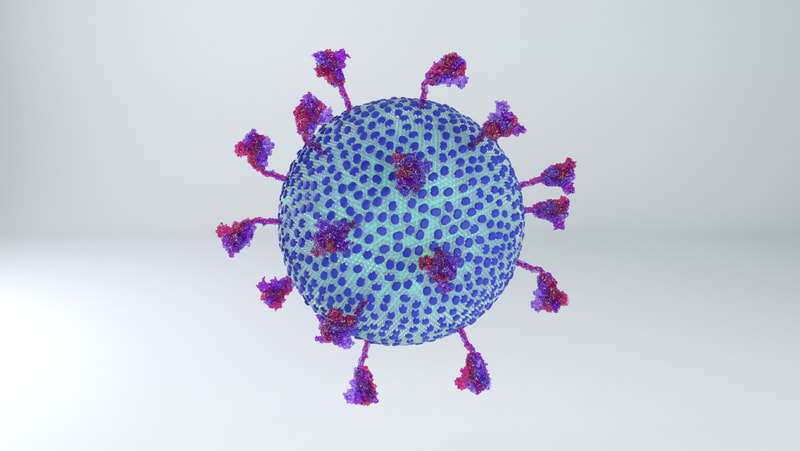
The most obvious change is the lower number of spikes. It's now known that the virus has an average of 24 spikes, and the spike proteins exist in a mixture of prefusion and postfusion shapes. The majority of spikes will be in the prefusion state, which is the form I've used for the spikes on the current model.
The height of the ectodomain region of each spike is at an average of 22.5 nm. The spikes rotate and travel randomly around the viral envelope. The spikes also flex, tilting at degrees of 50-90° in relation to the membrane.
Excluding spikes, the envelope sphere should be 88 nm on average in length. I've left it as a sphere, but it should be more of an ellipsoid, morphing blobbily from one contortion to another.
See the new video below:
The most obvious change is the lower number of spikes. It's now known that the virus has an average of 24 spikes, and the spike proteins exist in a mixture of prefusion and postfusion shapes. The majority of spikes will be in the prefusion state, which is the form I've used for the spikes on the current model.
The height of the ectodomain region of each spike is at an average of 22.5 nm. The spikes rotate and travel randomly around the viral envelope. The spikes also flex, tilting at degrees of 50-90° in relation to the membrane.
Excluding spikes, the envelope sphere should be 88 nm on average in length. I've left it as a sphere, but it should be more of an ellipsoid, morphing blobbily from one contortion to another.
See the new video below: